CANBIO - PhD Training Program
Individual projects - Intrinsic Escape Mechanisms
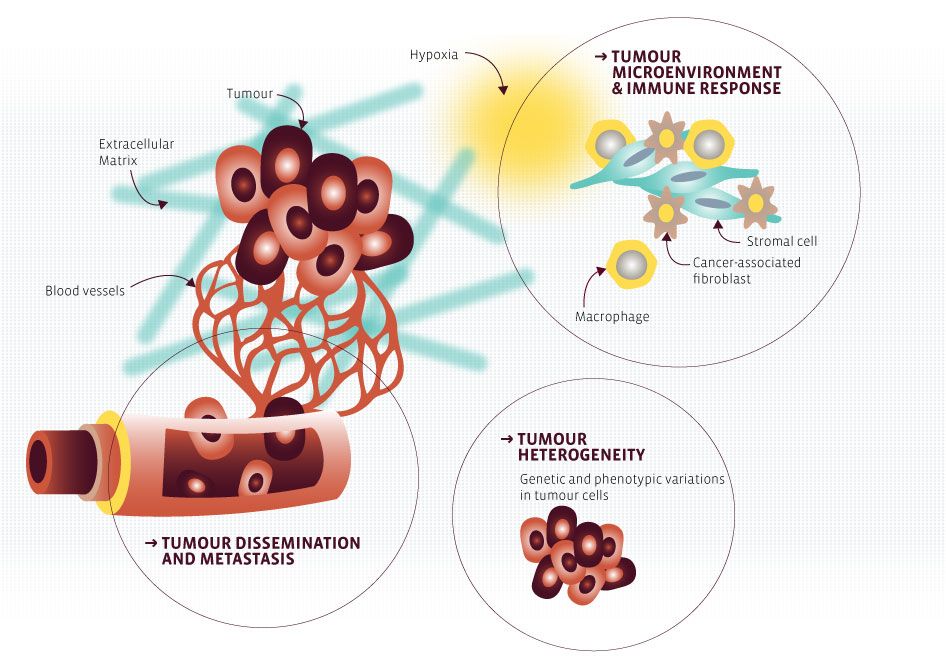
The first work package centres on the fundamental cancer question related to inherent tumour escape mechanisms: it will address the molecular aspects of tumour heterogeneity and the complex interplay between the tumour and its microenvironment, in particular with regard to immune cells
Such understanding aims at targeting cancer at its root and allows the exploitation of this new knowledge for the improvement of cancer therapies.
Project 1: Elucidating the genetics of invasive and tumour core compartments in glioblastoma
- Supervision : Prof Rolf Bjerkvig (LIH/UiB), Prof Simone Niclou (LIH)
Glioblastoma are composed of a dense tumour mass and of a highly invasive tumour margin, which infiltrates the brain parenchyma. We have recapitulated this growth behaviour in clinically-relevant animal models and have shown that tumours can switch to a more invasive growth in a treatment-dependent manner. Here we will analyse the underlying genetic and molecular mechanisms that are responsible for these patterns. This project will point to novel molecular pathways that should enable targeting the invasive component of Glioblastoma.
Project 2: The metabolic landscape of cancer-associated fibroblasts and its role in colorectal cancer progression and resistance
- Supervision: Dr Elisabeth Letellier (UL), Prof Serge Haan (UL),
Cancer-associated fibroblasts (CAFs) have been suggested to play an important role in tumor development, especially in relation to tumor initiation and metastasis. CAFs are highly heterogeneous and enhance cellular migration of epithelial tumor cells, display elevated pro-angiogenic cytokine signaling, and facilitate inflammation. They can also affect immune-recognition of tumor cells. By generating and integrating multi-omics data (transcriptomics, proteomics and metabolomics) of normal fibroblasts and tumor-associated fibroblasts from the same patient, we aim at building CAF-specific metabolic models and identify CAF-specific targets by using an integrated computational approach. Ultimately, this project aims at unraveling resistance mechanisms and new therapeutic strategies for colorectal cancer that are focused on the tumor microenvironment.
Project 3: The non-coding genome: contribution to tumour heterogeneity and functional implications in tumour development and drug resistance
- Supervision: Dr Stephanie Kreis (UL)
In this project, the focus will be on computationally exploring the non-coding (nc) parts of the genomes of melanoma: mutations and rearrangements (types and numbers) affecting promoters and other regulatory regions as well as miRNAs and lnc RNAs. The potential functional impact of the nc genome on tumour development and drug resistance will be explored in silico (RNA-Seq data generated from suitable cell models) followed by selected wet lab validation.
Project 4: Role of hypoxia in the establishment of an immune suppressive tumour microenvironment
- Supervision: Dr Bassam Janji (LIH) , Dr Guy Berchem (LIH)
Tumors use several mechanisms to evade the host immune control. It is now well established that hypoxia has not only a strong impact on tumor cell biology, but it also participates to the establishment of an immune suppressive tumor microenvironment. This project aims to understand the molecular mechanism(s) by which hypoxia-associated pathways drive tumor cell escape from immune cell attack and define innovative strategies to switch the immosuppressive to an immunosupportive environment in order to develop more durable and successful cancer immunotherapy approaches.
Project 5: Addressing the role of tumour associated microglia/macrophages in glioblastoma
- Supervision: Dr Alessandro Michelucci (LIH) , Prof Simone Niclou (LIH)
A major contributing factor to tumor development and progression is its ability to evade the immune system. Glioblastomas release factors that recruit resident microglia, macrophages and other peripheral immune cells to the tumor site and transform them into tumour-supportive cells. We aim to investigate the contribution of microglia/macrophages in intrinsic tumor escape mechanisms by analysing their phenotypic and functional adaptation in the tumorigenic process in order to reprogram them to promote anti-tumour activities.
Project 6: Identification of cytoskeleton targets to reverse immune resistance in breast cancer
- Supervision : Dr Clément Thomas (LIH) , Prof Michel Mittelbronn (LNS)
We recently discovered that breast cancer cells resist natural killer (NK) cell-induced cell death through a fast and prominent accumulation of actin at the cell-cell contact region. Our data indicate that such an actin-based response significantly reduces the delivery of cytolytic compounds, such as perforin and granzyme B, to the target cancer cells. The present project aims to further characterize the mechanism by which this actin response leads to immune
resistance. In addition, it aims to identify the cytoskeletal regulators driving the actin response, as well as the upstream signaling pathways. The use of such regulators and/or pathways as targets to reverse breast cancer cell resistance to natural killer cells will be evaluated.
DISCOVER CANBIO
About CANBIO - PhD Training Program
- WP1: Intrinsic Escape Mechanisms
- WP2: Induced Escape Mechanisms
- WP3: Disease Monitoring and Biological Networks
LuxDoc 3MT (Three Minute Thesis)
CONTACT
For any question related to CANBIO, please contact:
Prof. Simone Niclou
canbio.office@lih.lu
Funded by