CANBIO - PhD Training Program
Individual projects - Disease Monitoring and Biological Networks
New methods for the systematic stratification of patients and their treatment responses are needed to understand and overcome tumour escape mechanisms, be they intrinsic or acquired.
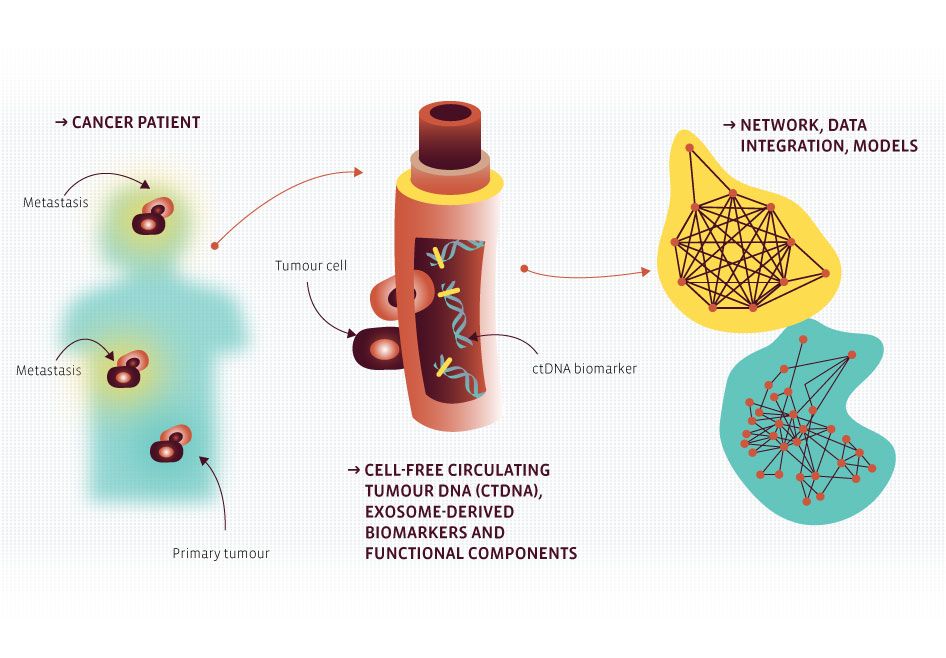
Within the third work package, novel approaches and methodologies to investigate biomarkers to monitor tumour escape mechanisms will be established, with a focus on liquid biopsies. The refined detection of new, specific and sensitive biomarkers alongside integrative systems-level data analysis will be applied to model and eventually predict disease progression and response to therapy. This knowledge will provide the basis for new discoveries and technical applications.
Project 1: Circulating tumour-derived DNA and miRNAs: cancer biomarkers in melanoma and other solid cancers
- Supervision: Dr Stephanie Kreis (UL)
Cell-free tumour DNA and also miRNAs are interesting molecules that can be indicative of disease progression and/or development of drug resistance. The project will analyse both molecule classes in melanoma patient-derived samples that are followed up during kinase inhibitor or immune therapy (if available) treatment with the aim to identify biomarkers that predict progression, response to treatment and developing resistance so that treatment regimens can be changed in time. Isolation and profiling of both cf-DNA and -miRNAs followed by bioinformatic analysis and validation will be core of the project.
Project 2: Exosome proteins and RNA as biomarkers of hematologic malignancies development and progression
- Supervision: Dr Bassam Janji (LIH/UPS) , Dr Etienne Moussay (LIH)
Tumor cells secrete small vesicles named exosomes which are involved in tumor stroma communication as well as in disease development and progression. Tumor-derived exosomes harbour a variety of proteins, nucleic acids, and lipids and are present in all body fluids. Exosomes draw much attention as a promising biomarker for tumor diagnosis and prognosis. This project aims to identify potential biomarkers in exosomes derived from murine preclinical models and cells of patients affected by hematologic malignancies and to test their influence on cells from the microenvironment. Candidates showing the greatest potential to be used in personalized medicine will be validated in cohorts of patients.
Project 3: The exosome proteome as a source of non-invasive biomarkers in solid tumours
- Supervision: Dr Gunnar Dittmar (LIH), Dr Guy Berchem (LIH/CHL)
Combining the advantages of non-invasive sampling and the availability of somatic variations, liquid biopsies using exosomes purified from patient blood are increasingly investigated as an alternative diagnostic solution. In this project, the student will develop LC-MS-based proteomic approach to analyse protein biomarkers of solid tumors in plasma exosomes in comparison to tissue biopsies. In addition to the activating somatic mutations of cancer drivers, post-translational modifications will be explored as biomarkers as well.
Project 4: Development of methodologies for monitoring cancer progression and predicting treatment response
- Supervision: Prof Thomas Sauter (LIH), Prof Simone Niclou (LIH)
The project will develop new computational models for predicting treatment responses. This will be achieved through the integrated analysis of different types of “omics” datasets. Among the modelling strategies to be implemented, the successful candidate will generate and analyse biological association networks for patient stratification. The project will investigate different applications with an emphasis on glioblastoma research. It will involve strong cooperation with the Kocher’s Lab at Mayo Clinic in the USA.
Project 5: Exploitation of multi-omics data generation capability and systems-based modelling approaches to investigating tumour escape mechanisms
- Supervision: Prof Thomas Sauter (UL)
The overwhelming amount of data calls for efficient computational tools allowing for integration and disease (patient) specific network analysis. We will further develop our algorithms for metabolic network reconstruction and establish a workflow for drug target identification based on cancer specific metabolic network models. In collaboration with Professor D. Kulms, experimental dermatology group at the university hospital Dresden, we will validate this new methodology on a melanoma cell line panel
Project 6: Circulating tumor DNA: Assessing different molecular strategies to achieve ultrasensitive detection of acquired somatic alterations.
- Supervision : Prof Michel Mittelbronn (LNS), Dr Daniel Stieber (LNS)
Cell-free tumor DNA derived from plasma (ctDNA) has shown potential as surrogate for tissue derived tumor DNA. Analysis of ctDNA can be applied to detect and monitor tumors in a non-invasive way, eliminating the need for repetitive invasive sampling and with the potential to comprehensively capture complete intra-tumoral genetic heterogeneity. To achieve this goal in a clinical diagnostic setting, a very high analytical sensitivity needs to be attained. The project aims to comparatively assess different ultrasensitive molecular strategies for their capacity to reliably detect recurrent molecular events in solid tumors from ctDNA. The clinical utility of this approach will be validated in a pilot study monitoring treatment response and disease progression in NSCLC patients.
DISCOVER CANBIO
About CANBIO - PhD Training Program
- WP1: Intrinsic Escape Mechanisms
- WP2: Induced Escape Mechanisms
- WP3: Disease Monitoring and Biological Networks
LuxDoc 3MT (Three Minute Thesis)
CONTACT
For any question related to CANBIO, please contact:
Prof Simone Niclou
canbio.office@lih.lu
Funded by