Grasping the regenerative capacity of the zebrafish heart at a systems level
The zebrafish is a key model system for vertebrate tissue regeneration after injury. Researchers from the NorLux Neuro-Oncology Laboratory at LIH’s Department of Oncology, together with other cross-disciplinary partners, have reported a study that offers novel insights into heart regeneration in the zebrafish based on the analysis of the underlying dynamic co-expression network. The findings were published in the journal Scientific Reports of the Nature Publishing group on 31st May 2016.
In mammals, the heart has a limited potential for regeneration after injury. The ability of organ and limb regeneration in lower vertebrate species is a fascinating research topic, essentially because the comprehension of the underlying molecular mechanisms could lead to the development of strategies to stimulate human organ regeneration. Since the 1970’s, the zebrafish has been a widely used genetic model organism to study how tissue regeneration occurs. These animals are capable of regenerating amputated fins and certain injured organs such as the brain, the spinal cord or the heart within weeks after damage. In the injured heart, proliferation is mainly triggered in existing cardiomyocytes that dedifferentiate and reactivate a cell developmental programme.
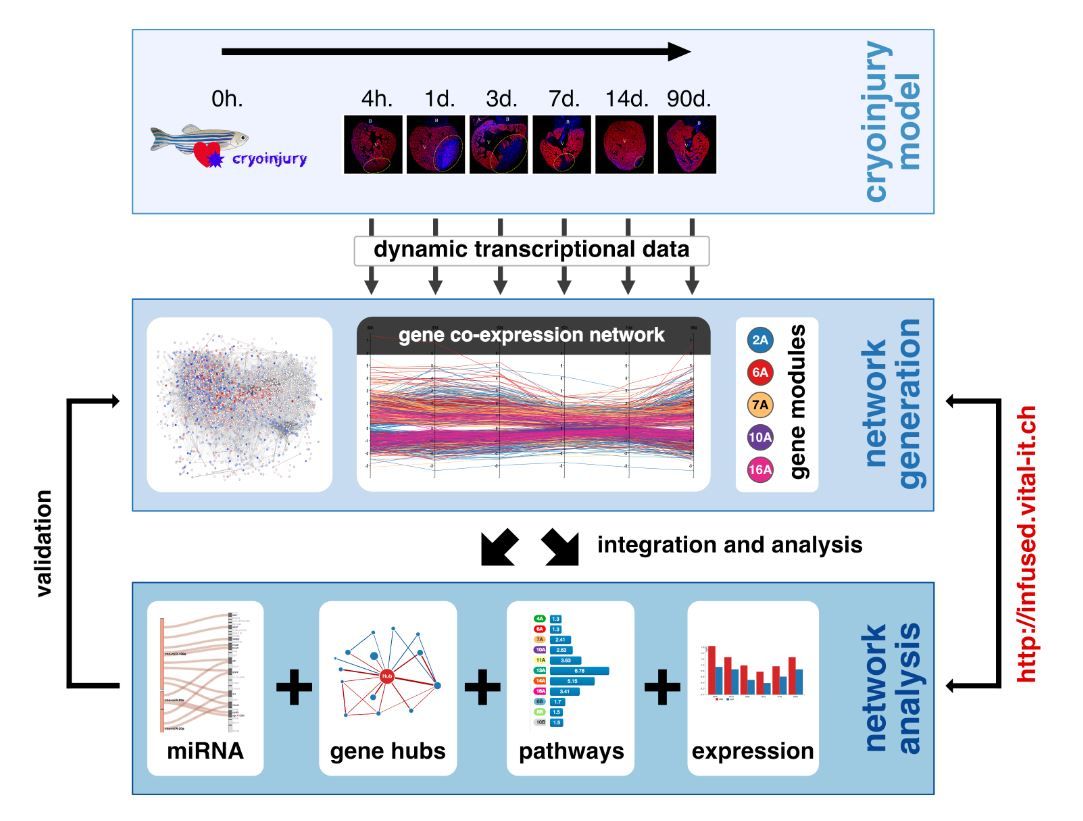
To get systematic insights into the phenomenon, the researchers performed transcriptional profiling of injured and control zebrafish hearts at different time-points after injury. They identified important time-specific changes in gene expression during heart generation, and generated a dynamic co-expression network consisting of thousands of genes and associations. The analysis of this network uncovered modules of highly interconnected genes, which are specialised in biological processes essential to heart regeneration. Furthermore, they identified hundreds of network hubs with control roles in heart regeneration. Interestingly, most of these hubs have human orthologs that are also the targets of known drivers of heart regeneration in mammals.
‘This is the first in-depth characterisation of the transcriptional network of heart regeneration in the zebrafish, and it highlights the value of systematic unbiased approaches to unravelling novel biological knowledge’, stresses Dr Francisco Azuaje, last author of the publication. ‘Another major outcome of our study is a web-based resource that allows the interactive analysis of relevant data and models reported in this article.’ This resource will be valuable to the zebrafish, heart regeneration and systems biology communities as it will greatly facilitate the understanding of zebrafish heart generation and the potential translation of this knowledge into therapeutic applications. ‘In the long-term, research in this area may enable new treatments for patients who suffer heart attacks’, states Dr Azuaje. ‘The induction of heart generation may be feasible, for example, through the targeting of network hubs or modules that are functionally conserved in zebrafish and humans.’
The research was funded by the National Research Fund of Luxembourg (FNR) and the Swiss National Research Foundation, as part of the INFUSED project. It involved researchers from the NorLux Neuro-Oncology Laboratory and the Genomics and Proteomics Research Unit at LIH, the Swiss Institute of Bioinformatics (Lausanne, Switzerland), the Centro Nacional de Investigaciones Cardiovasculares Carlos III (Madrid, Spain) and other institutions in Europe and the USA
Link to publication: www.nature.com/articles/srep26822
Link to web resource: http://infused.vital-it.ch
Figure 1 from publication: Discovery and resource development framework
(figure under the terms of the Creative Commons Attribution 4.0 International License http://creativecommons.org/licenses/by/4.0 )
